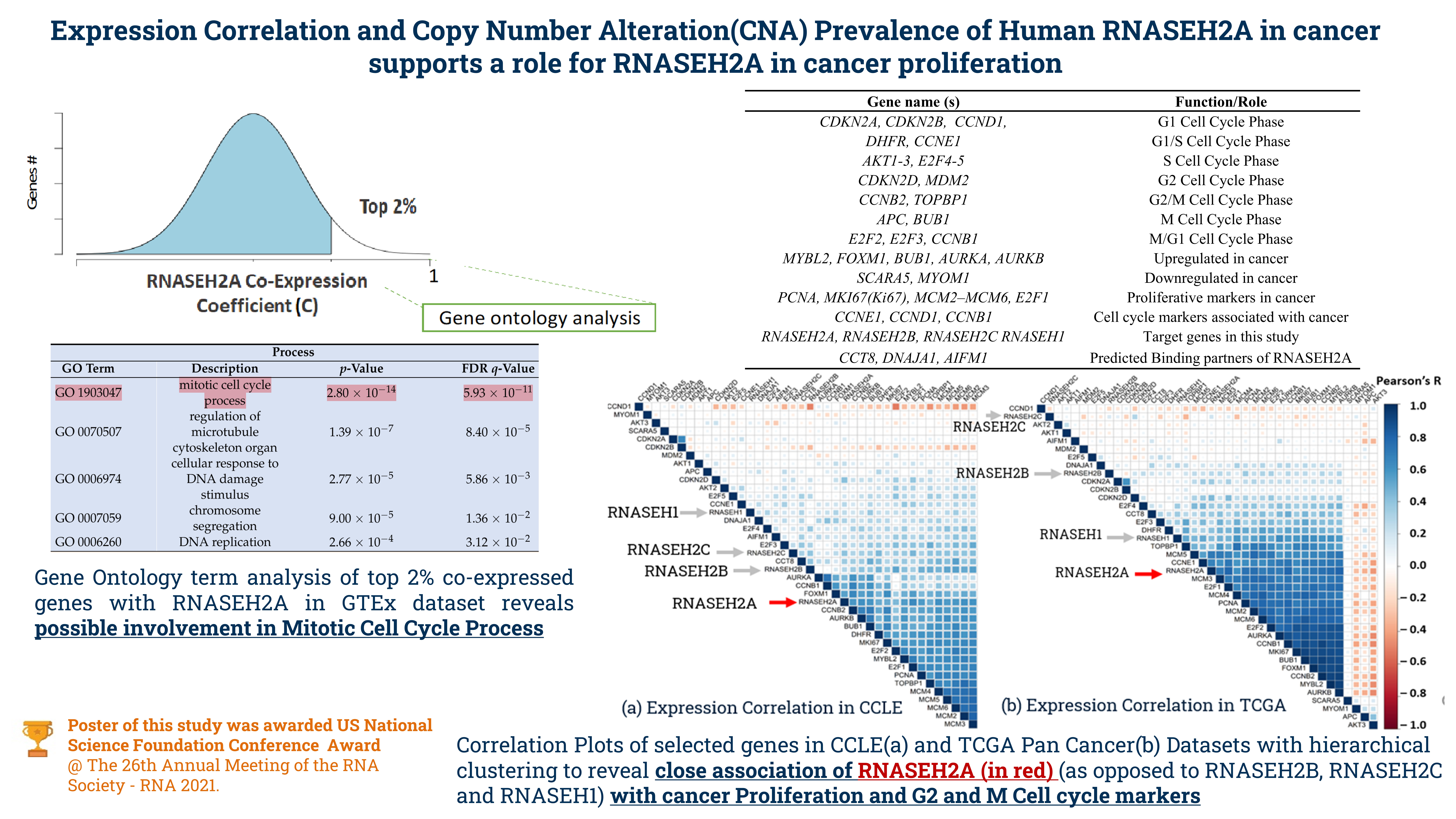
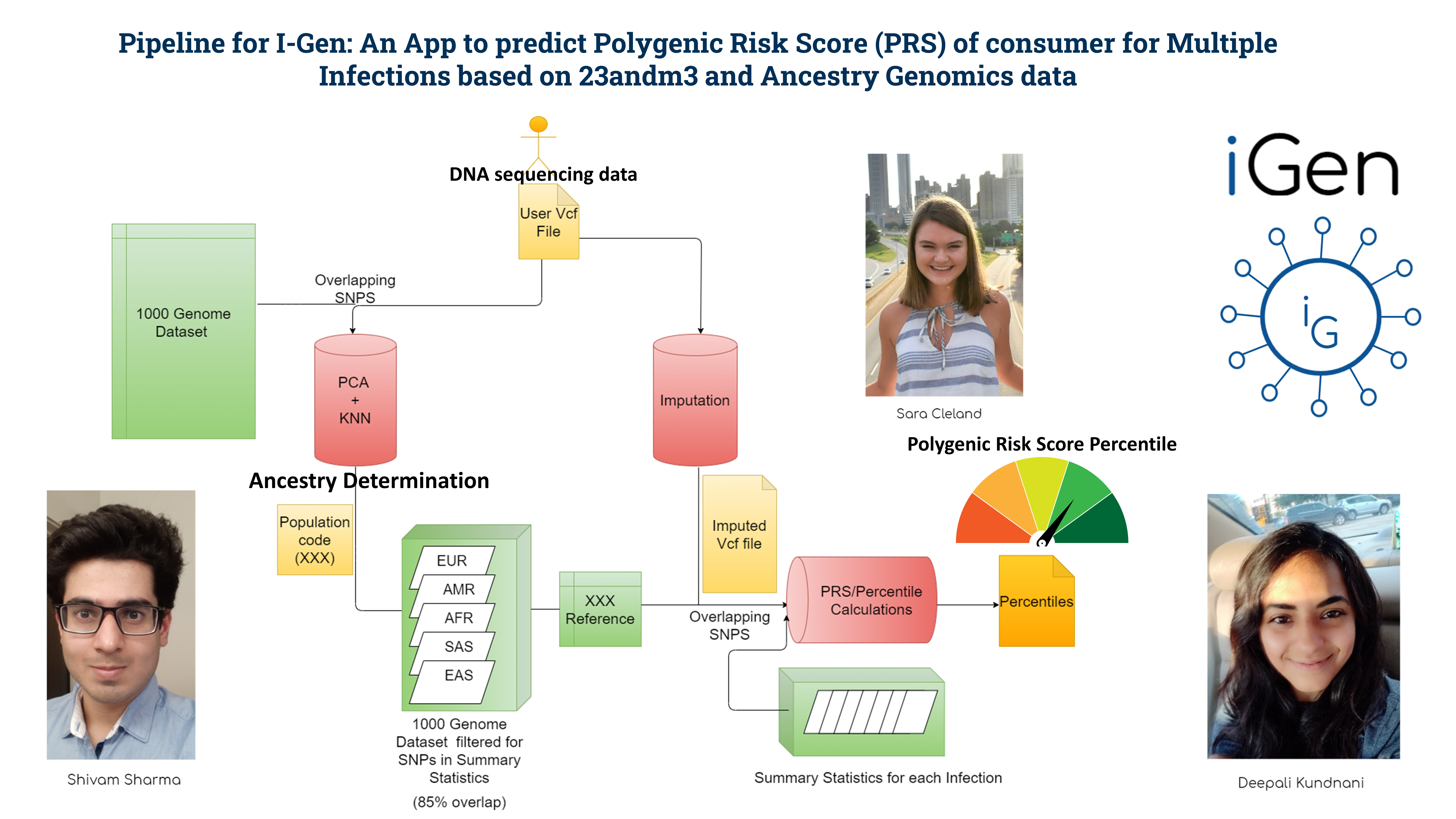
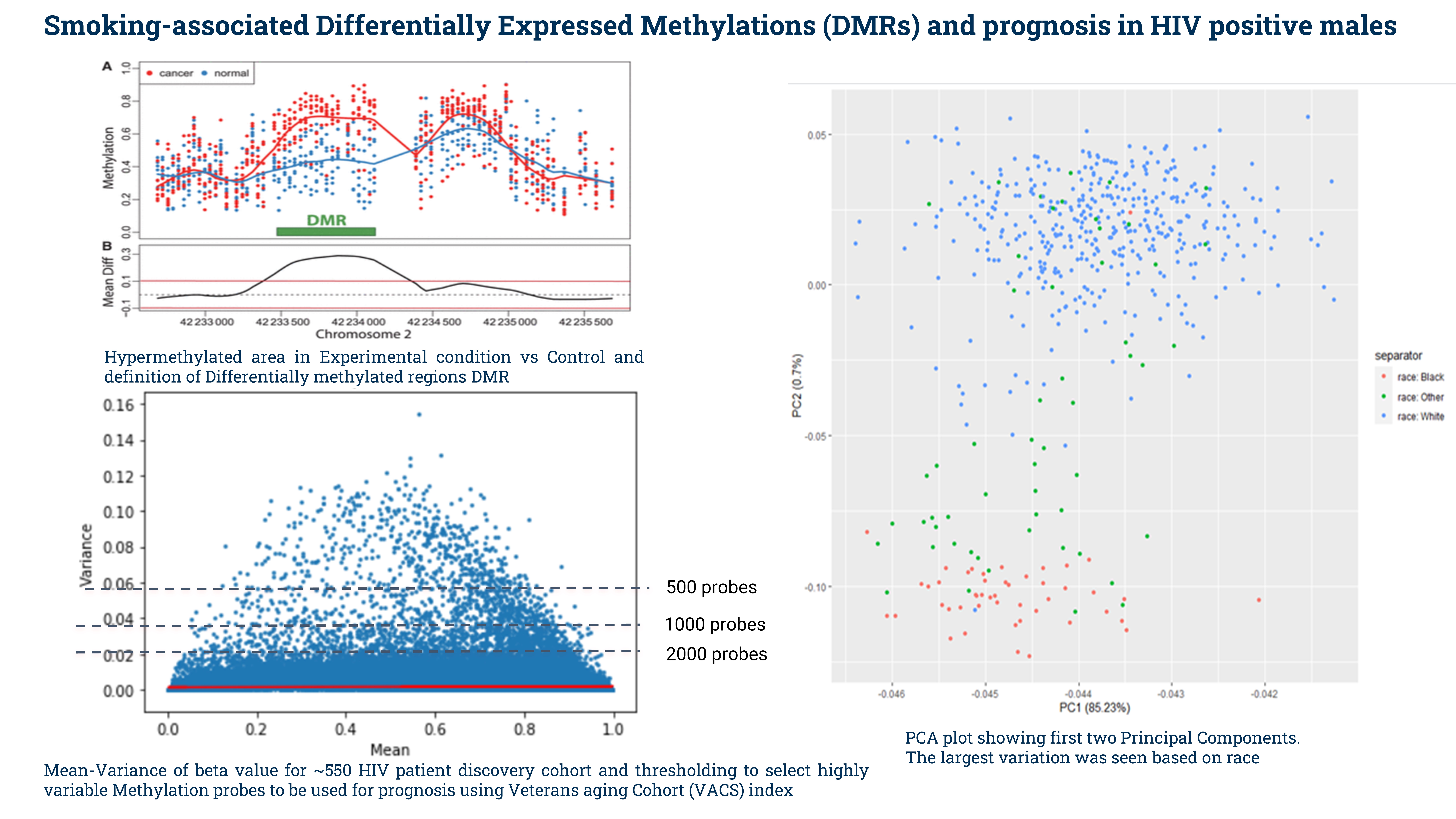
Recent Projects
Ribonucleotide enrichment features in human genome

Study of ribonucleotide embedment in human CD4+T cell type derived from
patient blood and lab grown stem cell line (hESC-H9) and embryonic
kidney cells (HEK293T), the latter with and without RNase H2 enzyme. In
this study we discovered single embedded ribonucleotides (rNMPs) are
enriched near CpG islands and are associated with gene expression and
methylation around Transcription Start Sites (TSSs). In addition, we see
a gene template vs non-template strand bias for RNase H2 KO that is not
present in wild-type RNase H2 cell associated with removal of rG on
non-template strand, possibly by Top1 enzyme.